☰
◀ Previous △ Index Next ▶
[Dicom]
 DICOM = Digital Imaging and Communications in Medicine. Is a
medical standard for distributing any kind of medical image.
flair has a capability to process the DICOM files with the use of
the pydicom module (available as an add-on package for your OS
distribution) and convert them to FLUKA VOXEL and/or USRBIN compatible
files. As well convert the RTPLAN and RTDOSE to a full input
for simulating a treatment planning.
The interface contains a tree browser on the left side that one
needs to fill with the appropriate DICOM files. The tree browser is
going to classify the files based on their modality and group them
with the same SOP UID class
DICOM = Digital Imaging and Communications in Medicine. Is a
medical standard for distributing any kind of medical image.
flair has a capability to process the DICOM files with the use of
the pydicom module (available as an add-on package for your OS
distribution) and convert them to FLUKA VOXEL and/or USRBIN compatible
files. As well convert the RTPLAN and RTDOSE to a full input
for simulating a treatment planning.
The interface contains a tree browser on the left side that one
needs to fill with the appropriate DICOM files. The tree browser is
going to classify the files based on their modality and group them
with the same SOP UID class
 Add will allow you to insert DICOM files to the project
Add will allow you to insert DICOM files to the project
 Will remove DICOM files from the project
Adding DICOM files in the project
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Click on the
Will remove DICOM files from the project
Adding DICOM files in the project
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Click on the  Add button.
The following dialog will allow you to select the files
Add button.
The following dialog will allow you to select the files
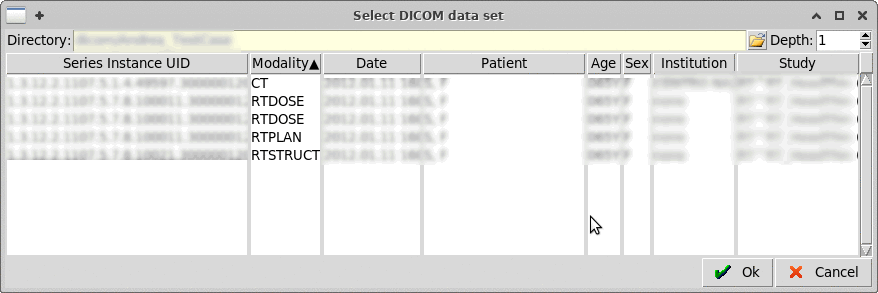 Select a parent directory containing the dicom files, up to a
"Depth" nesting of subfolders.
Warning: Do not increase a lot the depth as it can take considerable
amount of time to scan the sub-folders.
Dynamic Tabs
~~~~~~~~~~~~
Select a parent directory containing the dicom files, up to a
"Depth" nesting of subfolders.
Warning: Do not increase a lot the depth as it can take considerable
amount of time to scan the sub-folders.
Dynamic Tabs
~~~~~~~~~~~~
 Display information on the selected dataset
Dicom
Display information on the selected dataset
Dicom
 Show in a tabular text editor the dataset
Editor
Show in a tabular text editor the dataset
Editor
 Graphical dicom viewer
Viewer
Graphical dicom viewer
Viewer
 Convert a CT/NM/PT scan to a voxels file
Voxel
Convert a CT/NM/PT scan to a voxels file
Voxel
 Visualize and compare RTDOSE with USRBIN and their DVH
RTViewer
Visualize and compare RTDOSE with USRBIN and their DVH
RTViewer
 Convert an RTPLAN to input
RTPlan
Dicom Information
~~~~~~~~~~~~~~~~~
Convert an RTPLAN to input
RTPlan
Dicom Information
~~~~~~~~~~~~~~~~~
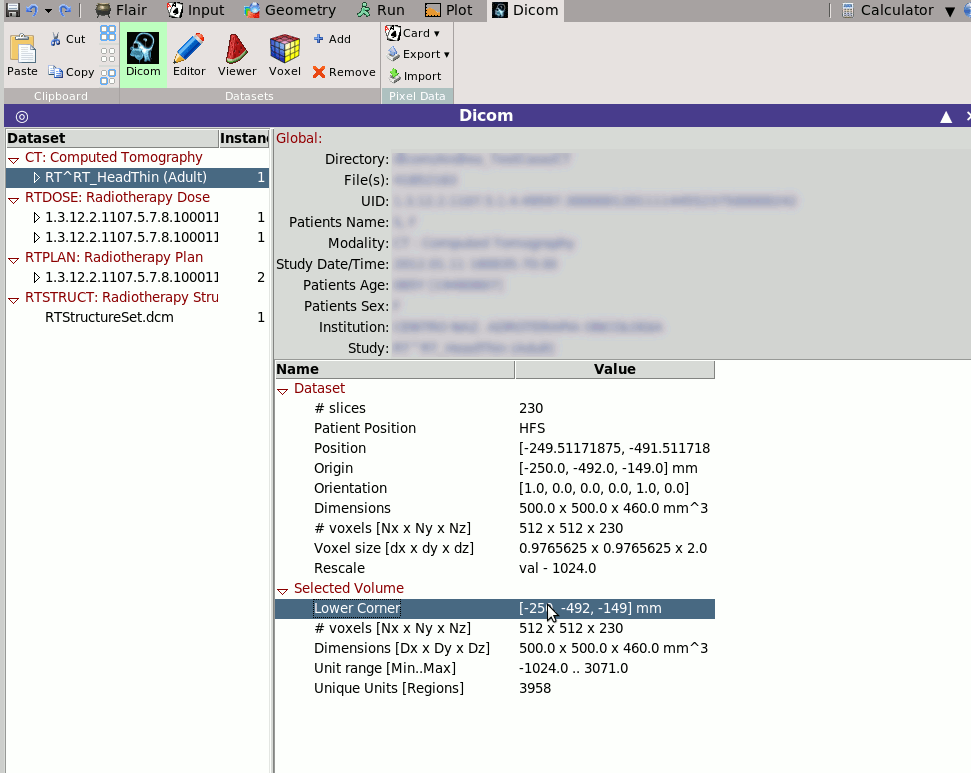 Selecting tab
Selecting tab  Dicom will display some generic
information on the selected dicom dataset.
The displayed information will depend on the selected modality.
e.g. when selecting an RTSTRUCT it will calculated and display all
ROIS and their respective volumes
Dicom will display some generic
information on the selected dicom dataset.
The displayed information will depend on the selected modality.
e.g. when selecting an RTSTRUCT it will calculated and display all
ROIS and their respective volumes
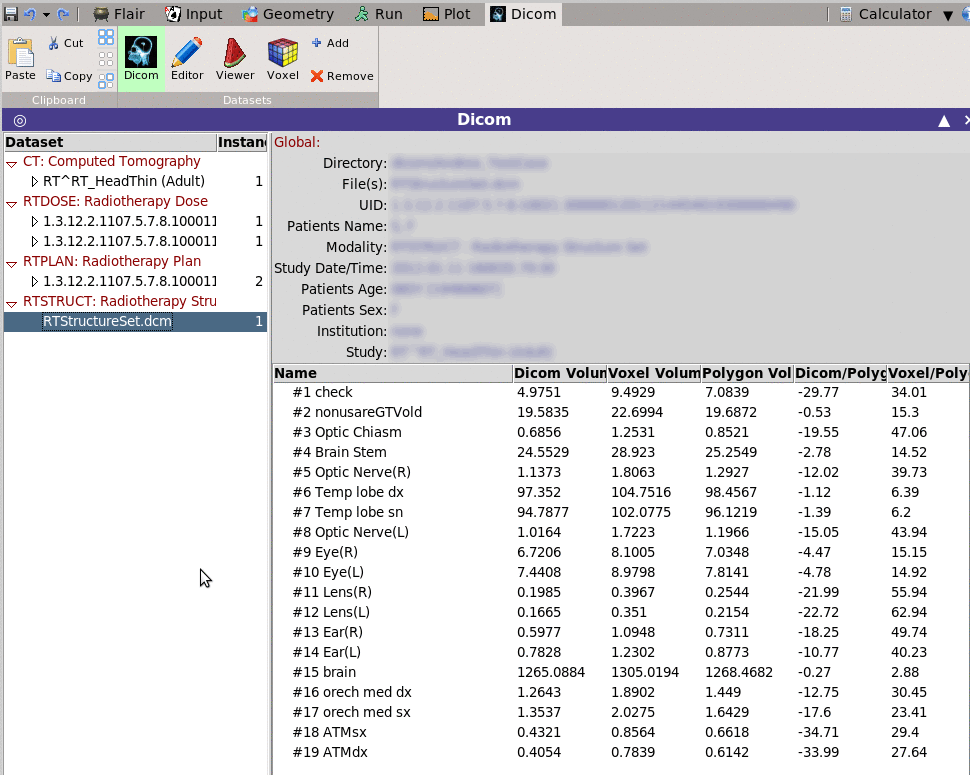 Dicom Editor
~~~~~~~~~~~~
Dicom Editor
~~~~~~~~~~~~
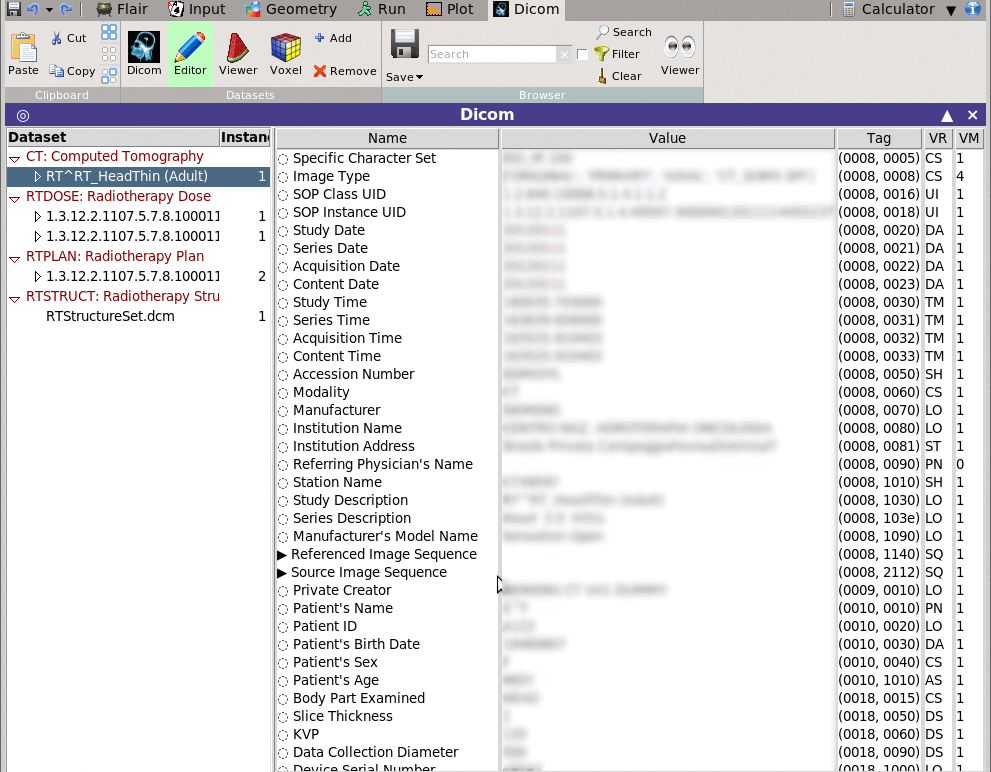 Shows in a multi-list tree as text the data stored in the selected dataset.
The Editor permits to change the value of the data and save the file.
For easier browsing you can filter and display only the items
matching a specific filter pattern.
Selecting multiple datasets (Shift+Right click) on the dataset
left-tree, the Editor will display the "common" entries of all
datasets.
Entries that are varied will be marked with a small dice-character
in-front.
With multiple selected files, changing an entry will change this
entry to ALL selected datasets.
Can be used to anonymize the selected datasets.
Slice Viewer
~~~~~~~~~~~~
Shows in a multi-list tree as text the data stored in the selected dataset.
The Editor permits to change the value of the data and save the file.
For easier browsing you can filter and display only the items
matching a specific filter pattern.
Selecting multiple datasets (Shift+Right click) on the dataset
left-tree, the Editor will display the "common" entries of all
datasets.
Entries that are varied will be marked with a small dice-character
in-front.
With multiple selected files, changing an entry will change this
entry to ALL selected datasets.
Can be used to anonymize the selected datasets.
Slice Viewer
~~~~~~~~~~~~
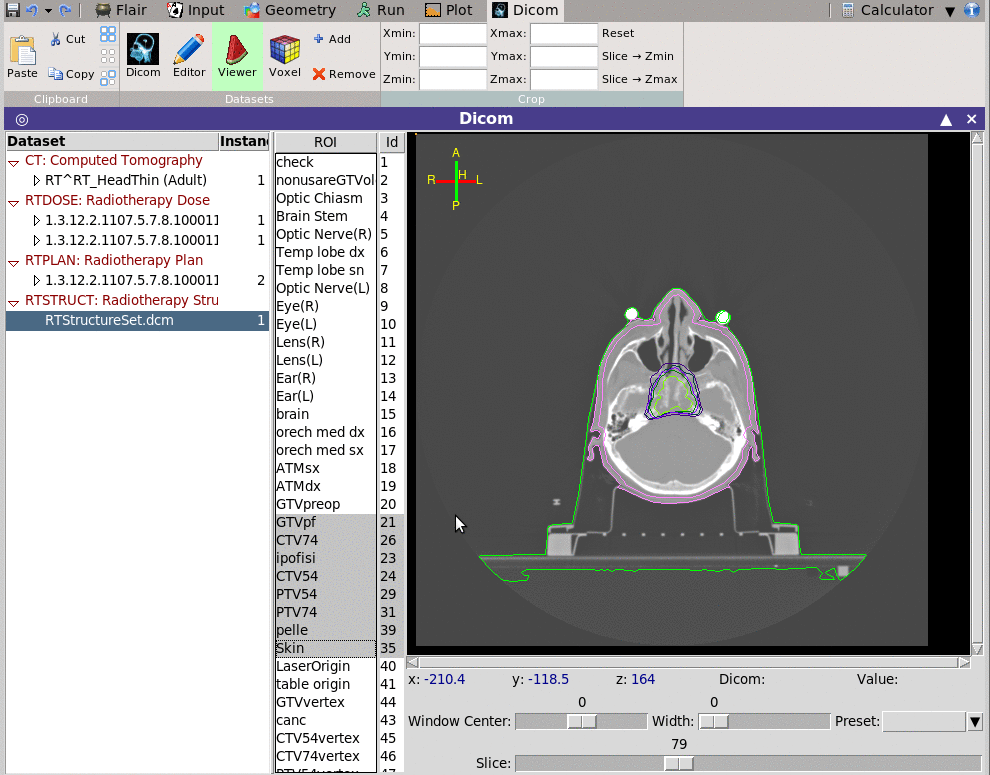 The viewer depending on the modality is able to show also the ROI
information.
Voxel Generation
~~~~~~~~~~~~~~~~
The viewer depending on the modality is able to show also the ROI
information.
Voxel Generation
~~~~~~~~~~~~~~~~
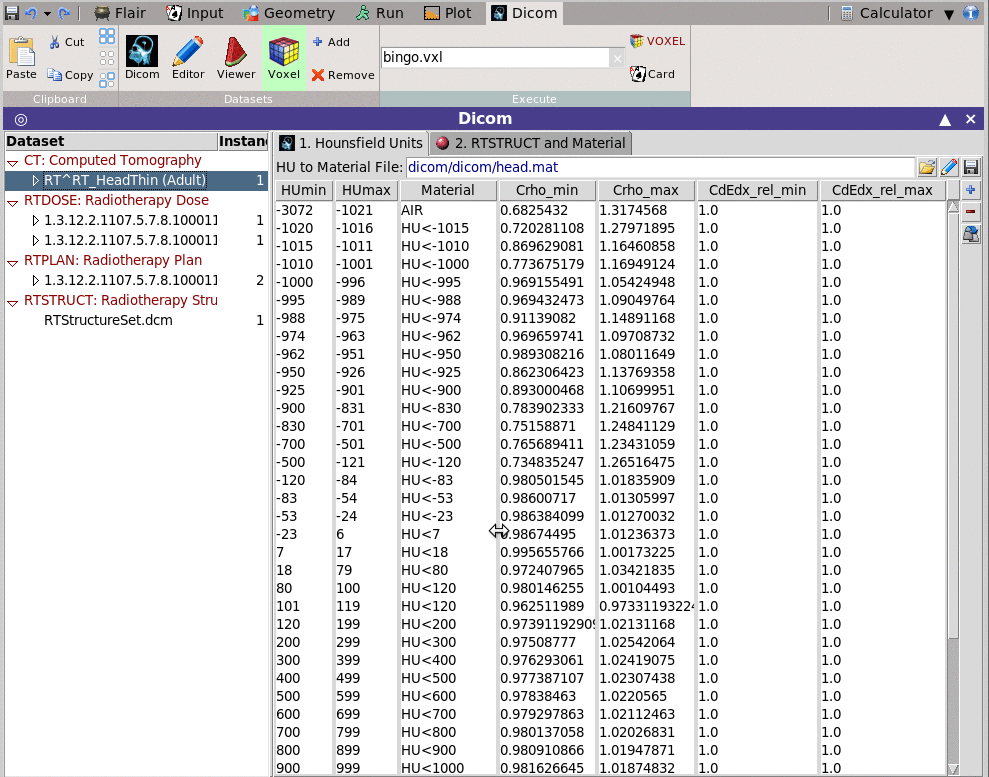 You can convert the CT scan and the associated RTSTRUCT to a voxel
format to be read by FLUKA
1. Hounsfield Units
~~~~~~~~~~~~~~~~~~~
You will need to provide a file containing the mapping mapping information
on how to convert the DICOM units (normally Hounsfield) to materials.
This can be done by editing the table below and save it to a file.
Click on <new> to add a new Units range
< Unit specify the upper limit (+1) of the range
Every entry will correspond to a range from the
previous upper limit until the current upper
limit-1
Material select any of the predefined FLUKA materials as
well the ones in the Materials input loaded before
Optionally you can specify correction factors for the density
and dE/dx
Crho_min density correction factor to be applied on the
lower limit of the unit range
Crho_max density correction factor to be applied on the
upper limit of the unit range
CdEdx_rel_min relative correction factor on dE/dx for the
minimum unit in the range
CdEdx_rel_max relative correction factor on dE/dx for the
maximum unit in the range
You can convert the CT scan and the associated RTSTRUCT to a voxel
format to be read by FLUKA
1. Hounsfield Units
~~~~~~~~~~~~~~~~~~~
You will need to provide a file containing the mapping mapping information
on how to convert the DICOM units (normally Hounsfield) to materials.
This can be done by editing the table below and save it to a file.
Click on <new> to add a new Units range
< Unit specify the upper limit (+1) of the range
Every entry will correspond to a range from the
previous upper limit until the current upper
limit-1
Material select any of the predefined FLUKA materials as
well the ones in the Materials input loaded before
Optionally you can specify correction factors for the density
and dE/dx
Crho_min density correction factor to be applied on the
lower limit of the unit range
Crho_max density correction factor to be applied on the
upper limit of the unit range
CdEdx_rel_min relative correction factor on dE/dx for the
minimum unit in the range
CdEdx_rel_max relative correction factor on dE/dx for the
maximum unit in the range
Notes:
~~~~~~
- The program will sort the Unit ranges in ascending order
- The first entry will correspond to all units from the
minimum in all slices to the upper limit defined by the user
- All units above the last entry will be assigned to the same
material as the last entry and a warning will be displayed
on the output window
- All intermediate units within a range will be scaled linearly
- dE/dx is applied relative to the correction of
density (negative WHAT(1)
in CORRFACT)
2. RTSTRUCT and Material
~~~~~~~~~~~~~~~~~~~~~~~~
This tab allows to override some structures with a fixed material
e.g. Everything outside the skin convert to Vacuum
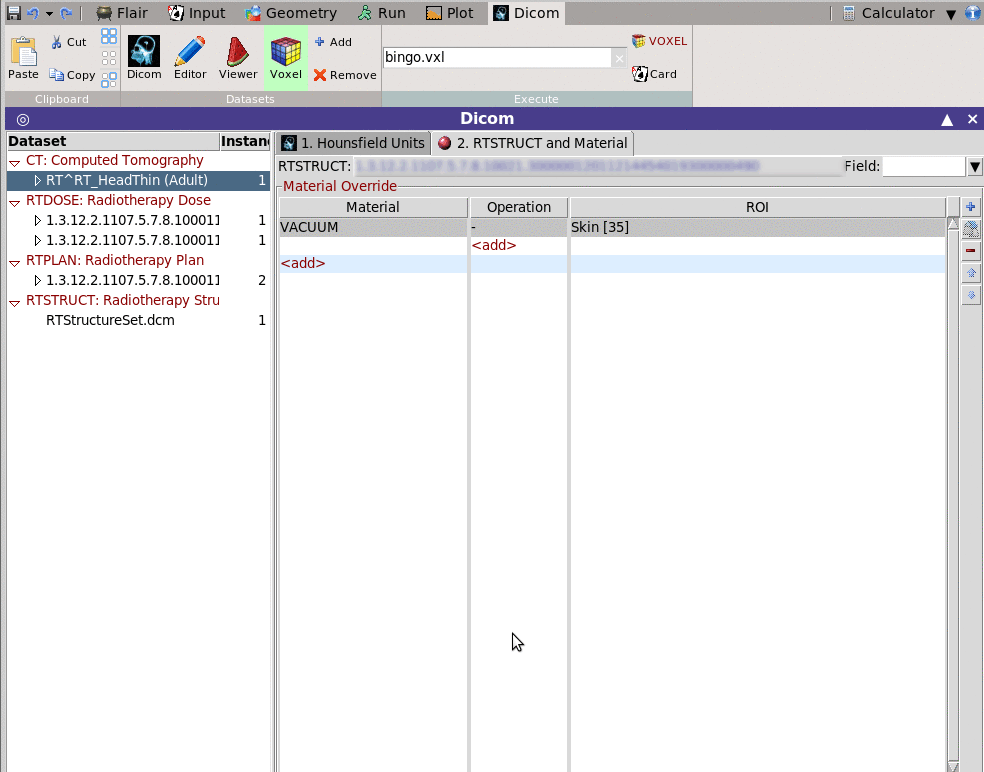 You need to fill in the Material override table a combinatorial
operation of the rois.
i. select the material override, e.g. VACUUM
ii. add pairs of operation operations and ROI
Operation can be:
+ inside of the ROI
- outside of the ROI
Buttons
-------
VOXEL Will create the voxel file
Card Will insert(or update) a VOXEL card in the input
with the correct coordinates of the VOXEL
RTPlan creation
~~~~~~~~~~~~~~~
You need to fill in the Material override table a combinatorial
operation of the rois.
i. select the material override, e.g. VACUUM
ii. add pairs of operation operations and ROI
Operation can be:
+ inside of the ROI
- outside of the ROI
Buttons
-------
VOXEL Will create the voxel file
Card Will insert(or update) a VOXEL card in the input
with the correct coordinates of the VOXEL
RTPlan creation
~~~~~~~~~~~~~~~
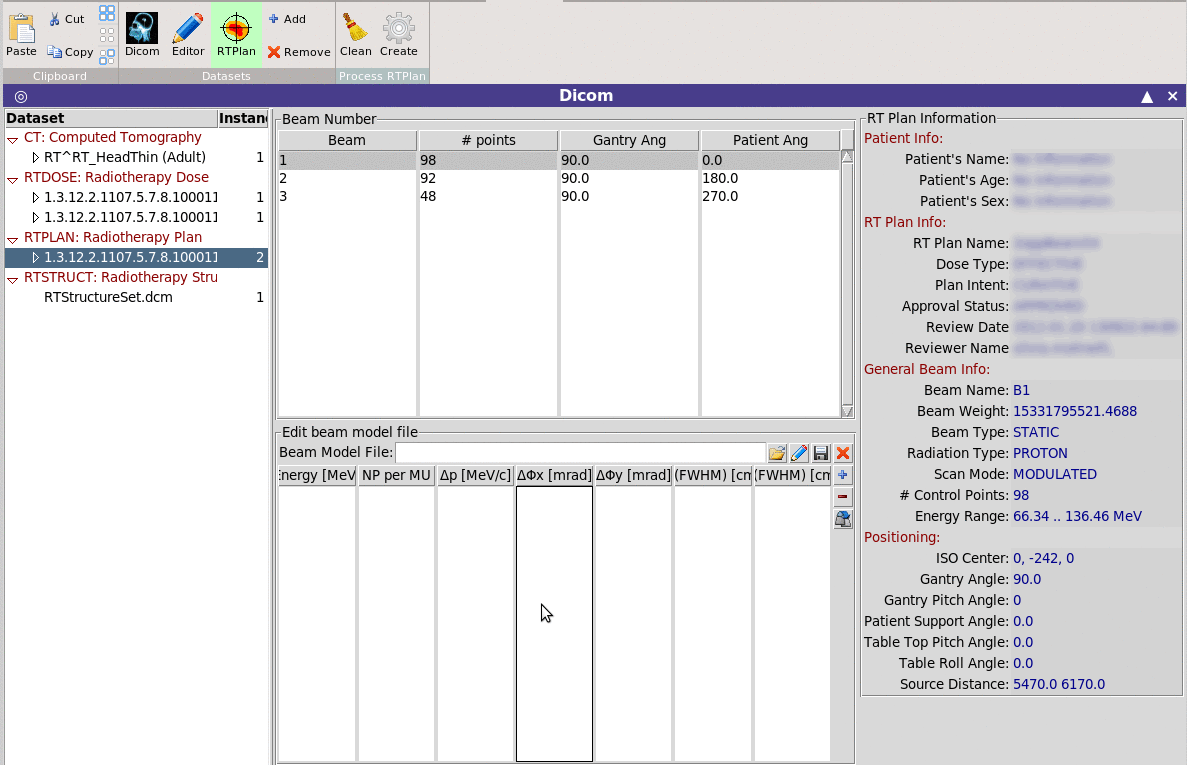 Allows the conversion of an RTPLAN dicom file to the equivalent
multi-spot beam input for FLUKA.
Limitations:
Currently it can create the files for one fraction and it does not
create the MLC structure in case of photon plans.
Beam model file:
Optionally you can provide a beam model file with additional
information on the beam, like momentum, angular and spatial spread
per beam energy.
The program will create a list of SPOTBEAM, SPOTPOS, SPOTDIR,
SPOTTRAN cards converted to the patient orientation system.
For each Beam it will create a separate #include file with all the
necessary cards.
The normalization for each beam will created with a special #define
variables that can be automatically used by the DVH converter and
RTViewer
The program will tag all cards generated with a special variable
in order to be able to delete and replace in case is needed.
RTViewer
~~~~~~~~
Allows the conversion of an RTPLAN dicom file to the equivalent
multi-spot beam input for FLUKA.
Limitations:
Currently it can create the files for one fraction and it does not
create the MLC structure in case of photon plans.
Beam model file:
Optionally you can provide a beam model file with additional
information on the beam, like momentum, angular and spatial spread
per beam energy.
The program will create a list of SPOTBEAM, SPOTPOS, SPOTDIR,
SPOTTRAN cards converted to the patient orientation system.
For each Beam it will create a separate #include file with all the
necessary cards.
The normalization for each beam will created with a special #define
variables that can be automatically used by the DVH converter and
RTViewer
The program will tag all cards generated with a special variable
in order to be able to delete and replace in case is needed.
RTViewer
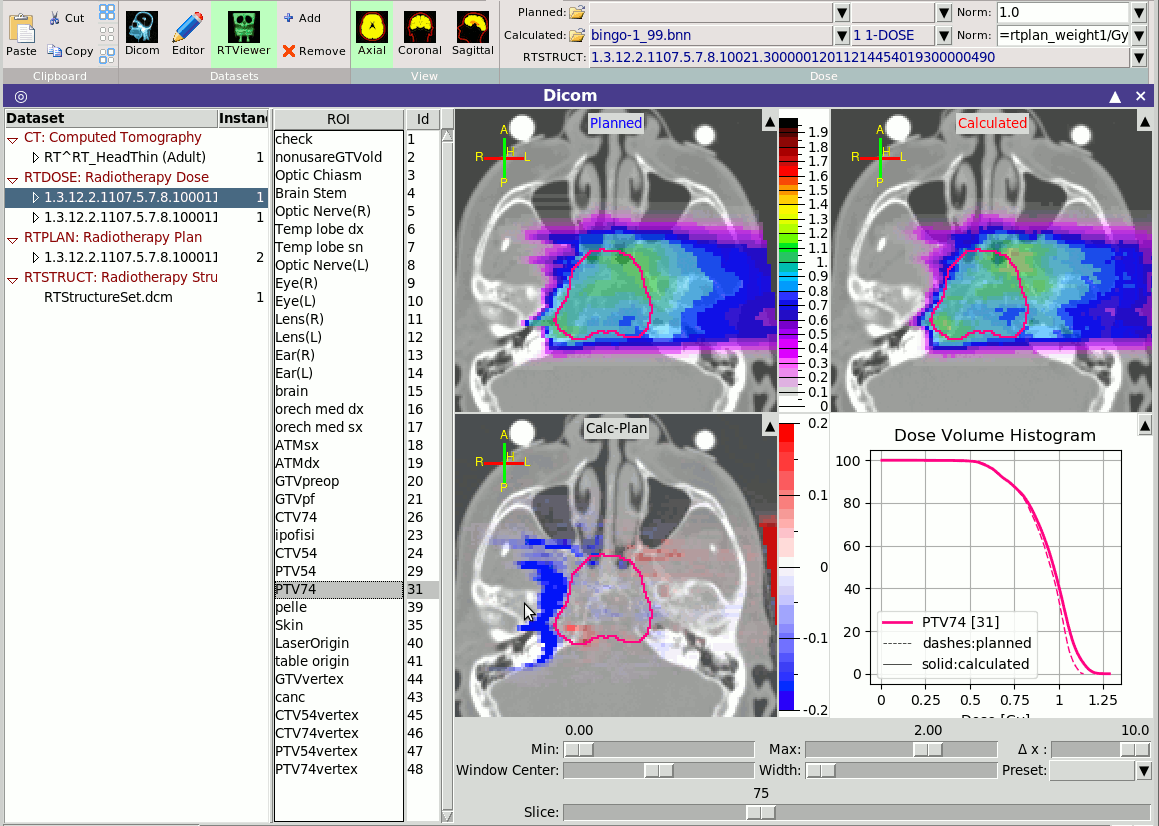
~~~~~~~~
 is a module linked with the RTDOSE modality to allow the comparison
of the planned dose vs the calculated dose from the MC.
You can load any RTDOSE and any USRBIN as planned or calculated,
provide the necessary normalization factor and compare the results.
When an RTDOSE or a USRBIN is loaded the program will try to
automatically create the DVH histograms, provided a voxels file is
present.
is a module linked with the RTDOSE modality to allow the comparison
of the planned dose vs the calculated dose from the MC.
You can load any RTDOSE and any USRBIN as planned or calculated,
provide the necessary normalization factor and compare the results.
When an RTDOSE or a USRBIN is loaded the program will try to
automatically create the DVH histograms, provided a voxels file is
present.
◀ Previous △ Index Next ▶
DICOM = Digital Imaging and Communications in Medicine. Is a medical standard for distributing any kind of medical image. flair has a capability to process the DICOM files with the use of the pydicom module (available as an add-on package for your OS distribution) and convert them to FLUKA VOXEL and/or USRBIN compatible files. As well convert the RTPLAN and RTDOSE to a full input for simulating a treatment planning. The interface contains a tree browser on the left side that one needs to fill with the appropriate DICOM files. The tree browser is going to classify the files based on their modality and group them with the same SOP UID class
Add will allow you to insert DICOM files to the project
Will remove DICOM files from the project Adding DICOM files in the project ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Click on the
Add button. The following dialog will allow you to select the files
Select a parent directory containing the dicom files, up to a "Depth" nesting of subfolders. Warning: Do not increase a lot the depth as it can take considerable amount of time to scan the sub-folders. Dynamic Tabs ~~~~~~~~~~~~
Display information on the selected dataset Dicom
Show in a tabular text editor the dataset Editor
Graphical dicom viewer Viewer
Convert a CT/NM/PT scan to a voxels file Voxel
Visualize and compare RTDOSE with USRBIN and their DVH RTViewer
Convert an RTPLAN to input RTPlan Dicom Information ~~~~~~~~~~~~~~~~~
Selecting tab
Dicom will display some generic information on the selected dicom dataset. The displayed information will depend on the selected modality. e.g. when selecting an RTSTRUCT it will calculated and display all ROIS and their respective volumes
Dicom Editor ~~~~~~~~~~~~
Shows in a multi-list tree as text the data stored in the selected dataset. The Editor permits to change the value of the data and save the file. For easier browsing you can filter and display only the items matching a specific filter pattern. Selecting multiple datasets (Shift+Right click) on the dataset left-tree, the Editor will display the "common" entries of all datasets. Entries that are varied will be marked with a small dice-character in-front. With multiple selected files, changing an entry will change this entry to ALL selected datasets. Can be used to anonymize the selected datasets. Slice Viewer ~~~~~~~~~~~~
The viewer depending on the modality is able to show also the ROI information. Voxel Generation ~~~~~~~~~~~~~~~~
You can convert the CT scan and the associated RTSTRUCT to a voxel format to be read by FLUKA 1. Hounsfield Units ~~~~~~~~~~~~~~~~~~~ You will need to provide a file containing the mapping mapping information on how to convert the DICOM units (normally Hounsfield) to materials. This can be done by editing the table below and save it to a file. Click on <new> to add a new Units range < Unit specify the upper limit (+1) of the range Every entry will correspond to a range from the previous upper limit until the current upper limit-1 Material select any of the predefined FLUKA materials as well the ones in the Materials input loaded before Optionally you can specify correction factors for the density and dE/dx Crho_min density correction factor to be applied on the lower limit of the unit range Crho_max density correction factor to be applied on the upper limit of the unit range CdEdx_rel_min relative correction factor on dE/dx for the minimum unit in the range CdEdx_rel_max relative correction factor on dE/dx for the maximum unit in the range
Notes:~~~~~~ - The program will sort the Unit ranges in ascending order - The first entry will correspond to all units from the minimum in all slices to the upper limit defined by the user - All units above the last entry will be assigned to the same material as the last entry and a warning will be displayed on the output window - All intermediate units within a range will be scaled linearly - dE/dx is applied relative to the correction of density (negativeWHAT(1)in CORRFACT) 2. RTSTRUCT and Material ~~~~~~~~~~~~~~~~~~~~~~~~ This tab allows to override some structures with a fixed material e.g. Everything outside the skin convert to VacuumYou need to fill in the Material override table a combinatorial operation of the rois. i. select the material override, e.g. VACUUM ii. add pairs of operation operations and ROI Operation can be: + inside of the ROI - outside of the ROI Buttons ------- VOXEL Will create the voxel file Card Will insert(or update) a VOXEL card in the input with the correct coordinates of the VOXEL RTPlan creation ~~~~~~~~~~~~~~~
Allows the conversion of an RTPLAN dicom file to the equivalent multi-spot beam input for FLUKA. Limitations: Currently it can create the files for one fraction and it does not create the MLC structure in case of photon plans. Beam model file: Optionally you can provide a beam model file with additional information on the beam, like momentum, angular and spatial spread per beam energy. The program will create a list of SPOTBEAM, SPOTPOS, SPOTDIR, SPOTTRAN cards converted to the patient orientation system. For each Beam it will create a separate #include file with all the necessary cards. The normalization for each beam will created with a special #define variables that can be automatically used by the DVH converter and RTViewer The program will tag all cards generated with a special variable in order to be able to delete and replace in case is needed. RTViewer ~~~~~~~~
is a module linked with the RTDOSE modality to allow the comparison of the planned dose vs the calculated dose from the MC. You can load any RTDOSE and any USRBIN as planned or calculated, provide the necessary normalization factor and compare the results. When an RTDOSE or a USRBIN is loaded the program will try to automatically create the DVH histograms, provided a voxels file is present.
 flair
flair